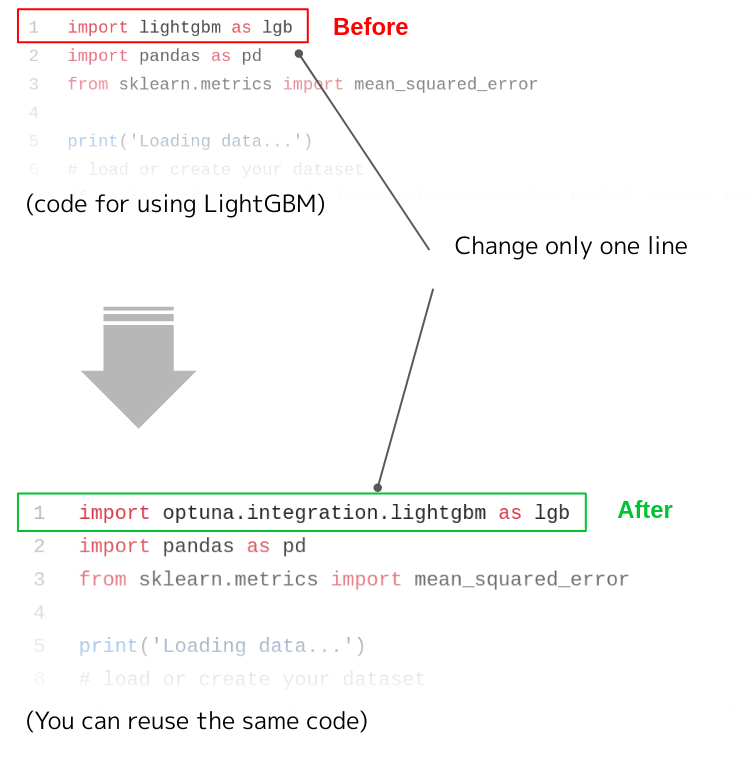
Blog
2020.01.29
Introduction to Kurobako: A Benchmark Tool for Hyperparameter Optimization Algorithms
Tag
Takeru Ohta
Engineer

“kurobako” is a Japanese translation of “black box” (image from iconmonstr.com)
Kurobako is a command-line benchmark tool for hyperparameter optimization algorithms. It is framework agnostic but mainly used in the development process of Optuna, a hyperparameter optimization framework, to verify the optimization performance.
In this article, I will briefly introduce this benchmark tool and describe why this is needed by Optuna.
Optuna and blackbox optimization
Optuna[1] is a hyperparameter optimization framework. Optuna consists of a sampler and pruner, which can be changed to customize the optimization algorithm.
The sampler decides which hyperparameter to optimize on the next trial aiming to maximize or minimize the objective function, which the user defines (Figure 1). It doesn’t require any special characteristic of the objective function (such as differentiability) and runs only based on past hyperparameter evaluations. This kind of optimization algorithm is called blackbox optimization.
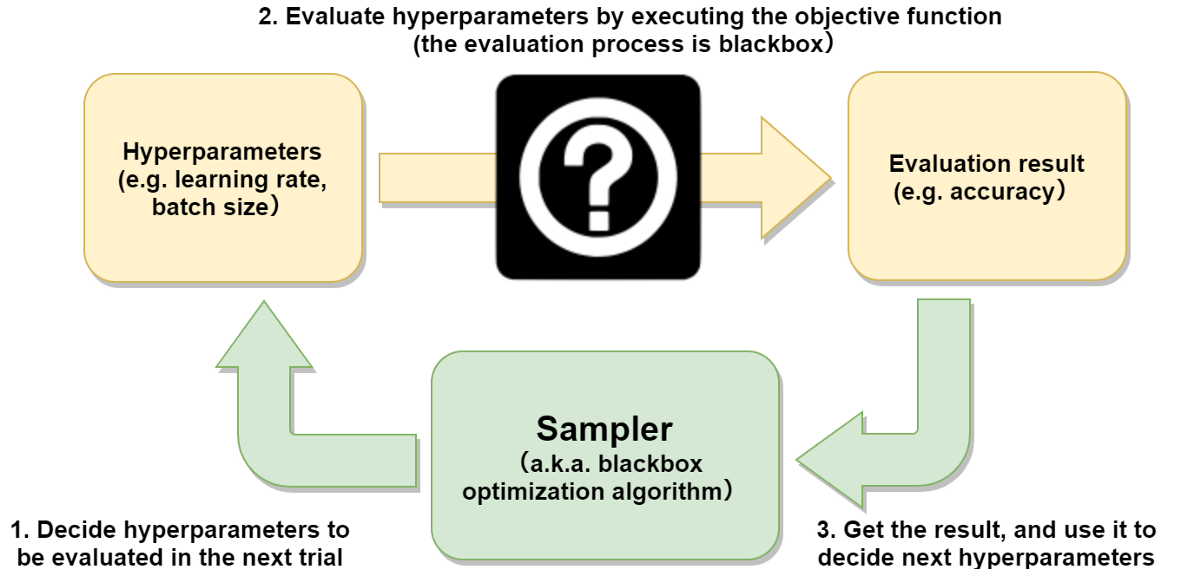
Figure 1: Blackbox optimization
On the other hand, the pruner will decide whether to discontinue the evaluation of the hyperparameter. In deep learning, suppose we want to optimize the hyperparameter to train the model, it often takes a long time for each evaluation of the hyperparameter. By using the pruner, we can shorten the time to optimize by aborting the evaluation(training) of models which are clearly not performing in the early phase and not completely evaluating them. In the context of blackbox optimization, the sampler and pruner are not clearly distinguished, and both are often addressed in the framework of multi-fidelity blackbox optimization (Figure 2).
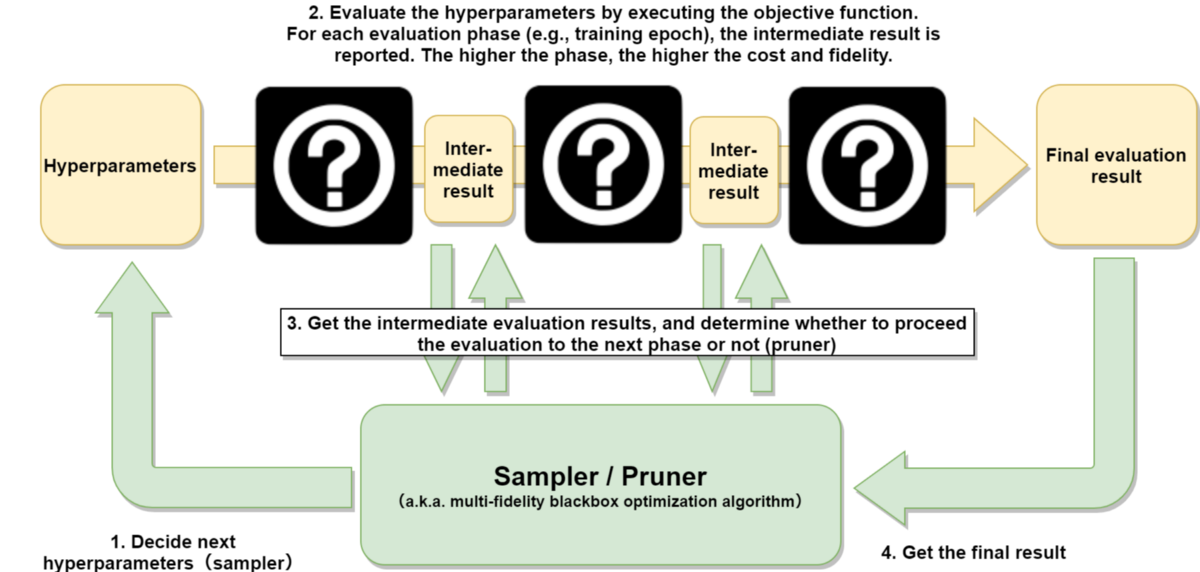
Figure 2: Multi-fidelity blackbox optimization
Kurobako is a benchmark tool that supports multi-fidelity blackbox optimization.
Motivation
Through development of Optuna, there are many occasions where the algorithm for sampler or pruner is tested or improved. The problem arises — “How to evaluate the optimization performance of new algorithms?”
Blackbox optimization is a research area that has a long history, and many benchmark functions that can be used to evaluate optimization algorithms have been developed[2]. One such open source benchmark framework is COCO (COmparing Continuous Optimisers)[3]. However, most of these functions can only deal with search space that consists of continuous variables and single fidelity, which is only a subset of the problems that Optuna addresses (Figure 3).
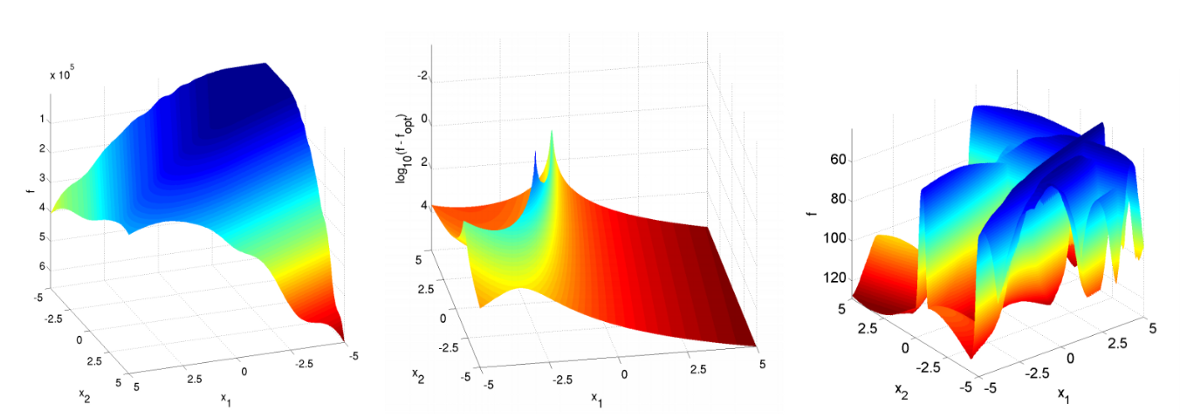
Figure 3: A part of the benchmark functions included in COCO (the full list)
A benchmark tool that deals with problems that are more realistic is nas_benchmarks. This provides benchmarks that are based on two tasks, Neural Architecture Search (NASBench)[4] and Hyperparameter Search (HPOBench)[5]. The search space consists of discrete variables and categorical variables (Figure 4). Also, it can deal with multi-fidelity evaluation.
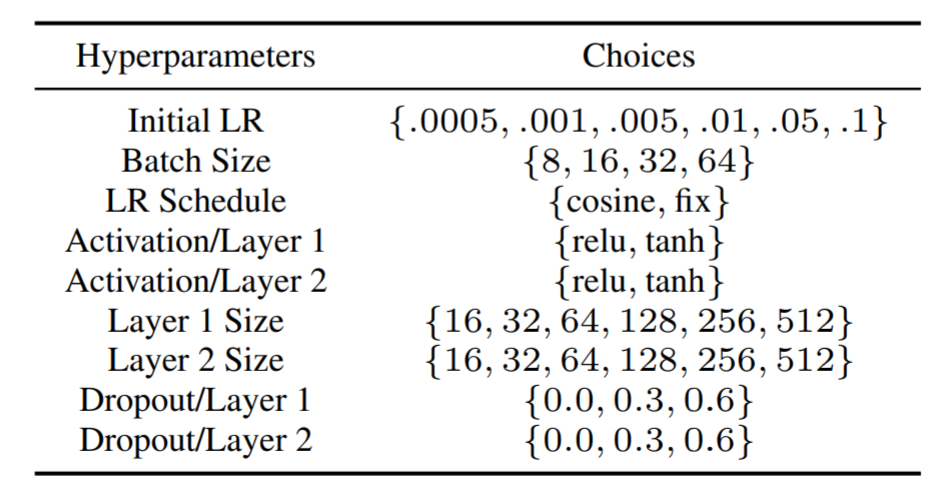
Figure 4: HPOBench search space[5]
The above benchmarks are useful but do not match our needs.
In particular, to evaluate the main optimization functions in Optuna, below must be supported:
- Search space including continuous, discrete, and categorical variables
- Multi-fidelity evaluation
- Asynchronous parallel execution of multiple trials
- Conditional search space including if and for loops
To the best of my knowledge, no benchmark tool can deal with all of the above. Motivation to develop Kurobako is to be able to cover all of this so that all optimization functions of Optuna can be evaluated easily.
Also, evaluating a certain benchmark can result in the optimization algorithm (and the hyperparameter) overfitting the benchmark. To prevent this, we wanted to be able to add a new benchmark problem easily and deal with various benchmark classes in the same framework.
Currently, in addition to NASBench and HPOBench, sigopt/evalset[6] benchmark functions are provided in Kurobako (version 0.1.4).
Basic usage
Kurobako is a command-line tool written in Rust, and uses JSON for input/output.
The following commands show the basic usage:
# 1. Download kurobako binary.
$ curl -L https://github.com/sile/kurobako/releases/download/0.1.4/kurobako-0.1.4.linux-amd64 -o kurobako
$ chmod +x kurobako && sudo mv kurobako /usr/local/bin/
# 2. Download the data file for HPOBench (note that the file size is about 700MB).
$ curl -OL http://ml4aad.org/wp-content/uploads/2019/01/fcnet_tabular_benchmarks.tar.gz
$ tar xf fcnet_tabular_benchmarks.tar.gz && cd fcnet_tabular_benchmarks/
# 3. Specify problems used in this benchmark.
#
# In this example, we use Protein Structure and Parkinsons Telemonitoring datasets.
$ kurobako problem hpobench fcnet_protein_structure_data.hdf5 > problems.json
$ kurobako problem hpobench fcnet_parkinsons_telemonitoring_data.hdf5 >> problems.json
$ cat problems.json
{"hpobench":{"dataset":"fcnet_protein_structure_data.hdf5"}}
{"hpobench":{"dataset":"fcnet_parkinsons_telemonitoring_data.hdf5"}}
# 4. Specify optimization algorithms ("solver" in Kurobako's terminology) used in this benchmark.
#
# In this example, we use the following four algorithms provided by Optuna:
# - RandomSampler without pruning
# - TPESampler without pruning
# - TPESampler with MedianPruner
# - TPESampler with SuccessiveHalvingPruner
$ kurobako solver --name 'RandomSampler' optuna --sampler random --pruner nop > solvers.json
$ kurobako solver --name 'TPESampler' optuna --sampler tpe --pruner nop >> solvers.json
$ kurobako solver --name 'TPESampler_with_MedianPruner' optuna --pruner median --median-warmup-steps 4 >> solvers.json
$ kurobako solver --name 'TPESampler_with_SuccessiveHalvingPruner' optuna --pruner asha --asha-min-resource 4 >> solvers.json
$ cat solvers.json
{"name":"RandomSampler","optuna":{"sampler":"random","pruner":"nop"}}
{"name":"TPESampler","optuna":{"pruner":"nop"}}
{"name":"TPESampler with MedianPruner","optuna":{"median_warmup_steps":4}}
{"name":"TPESampler with SuccessiveHalvingPruner","optuna":{"pruner":"asha","asha_min_resource":4}}
# 5. Run the benchmark.
$ kurobako studies --solvers $(cat solvers.json) --problems $(cat problems.json) --repeats 30 --budget 80 > studies.json
$ cat studies.json | kurobako run --parallelism 10 > result.json
(ALL) [01:27:14] [STUDIES 240/240 100%] [ETA 0s] done
# 6. Generate the report of the benchmark result.
$ cat result.json | kurobako report > report.md
You can find the content of the last report in the gist.
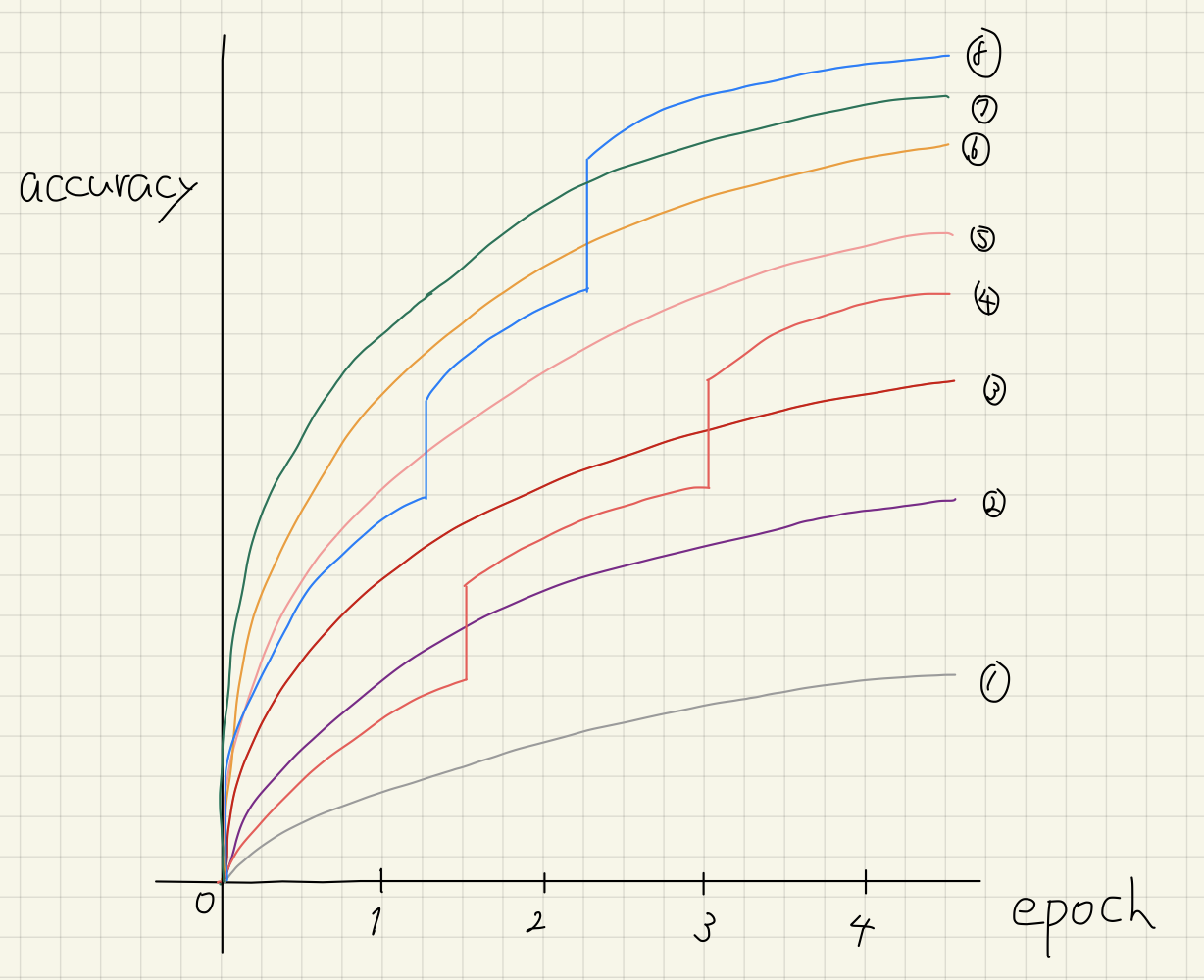
It is possible to generate figures from the benchmark result (result.json) by executing the command $ cat result.json | kurobako plot curve (Figure 5).
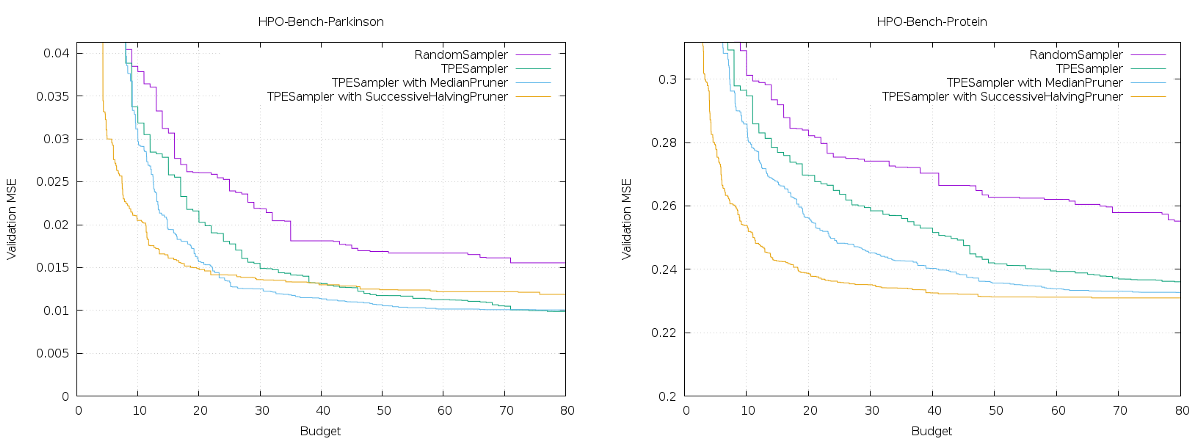
Figure 5: Optimization curves
Y axis in graph is the best results (average of 30 results). X axis shows resource consumption of optimization, 1 budget corresponds to one complete evaluation of objective function. In HPOBench, a complete evaluation (training of model) takes 100 epochs, so if the budget is 40, training for 4000 epochs have been run. In HPOBench, all hyperparameter combinations in the search space are evaluated beforehand, and the results are cached. So on benchmark runs, model training is unnecessary, and only cache lookup is necessary.
The benchmark above is only an example, but when the available budget is limited, enabling pruning will give better results. However, as the available budget increases, the effect of pruning diminishes. Especially in the last stage of Parkinsons Telemonitoring Data Set, TPE with pruning by Successive Halving algorithm[7] is worse than without pruning.
How to add optimization algorithm
Kurobako is written in Rust, but It is possible to implement an original optimization algorithm (solver) as a command line program. (As is for problem)
For Python, kurobako package is provided in PyPI, so solvers can be implemented as below:
# A solver implementation based on Random Search algorithm.
from kurobako import problem
from kurobako import solver
import numpy as np
class RandomSolverFactory(solver.SolverFactory):
def specification(self):
return solver.SolverSpec(name='Random Search')
def create_solver(self, seed, problem):
return RandomSolver(seed, problem)
class RandomSolver(solver.Solver):
def __init__(self, seed, problem):
self._rng = np.random.RandomState(seed)
self._problem = problem
def ask(self, idg):
trial_id = idg.generate()
next_step = self._problem.steps.last_step
params = []
for p in self._problem.params:
if p.distribution == problem.Distribution.LOG_UNIFORM:
low = np.log(p.range.low)
high = np.log(p.range.high)
params.append(float(np.exp(self._rng.uniform(log, high))))
else:
params.append(self._rng.uniform(p.range.low, p.range.high))
return solver.NextTrial(trial_id, params, next_step)
def tell(self, trial):
pass
if __name__ == '__main__':
runner = solver.SolverRunner(RandomSolverFactory())
runner.run()
If the optimization algorithm is compliant to Optuna’s sampler interface, it is easier:
# A solver implementation based on Simulated Annealing algorithm.
from kurobako import solver
from kurobako.solver.optuna import OptunaSolverFactory
import optuna
class SimulatedAnnealingSampler(optuna.BaseSampler):
# Please refer to
# https://github.com/optuna/optuna/blob/v1.0.0/examples/samplers/simulated_annealing_sampler.py
# for the implementation.
...
def create_study(seed):
sampler = SimulatedAnnealingSampler(seed=seed)
return optuna.create_study(sampler=sampler)
if __name__ == ‘__main__’:
factory = OptunaSolverFactory(create_study)
runner = solver.SolverRunner(factory)
runner.run()
You can specify the above solvers in your benchmark as follows:
$ kurobako solver command python random.py > solvers.json $ kurobako solver command python sa.py >> solvers.json $ kurobako studies --problems $(cat problems.json) --solvers $(cat solvers.json) | kurobako run > result.json
Conclusion
In this post, I introduced a benchmark tool for blackbox optimization. Please test this if you are interested in contributing to Optuna’s algorithm but are not sure about the optimization performance. Final goal is to integrate the benchmark to Optuna’s CI.
Though not discussed in this post, Kurobako supports features not supported in current Optuna, such as multi-objective optimization, optimization with constraint, suspend/resume of trials. If you use it in a special way, you can use it for meta optimization use. Examples are “hyperparameter optimization of the solver” (setting solver as the problem) or, even “searching for default parameters that perform well over multiple problems”.
If you are interested in Kurobako, please refer to the README or Wiki for more information.
Acknowledgment
This is an English translation version of my Japanese blog post. The translation was mainly done by Hiroshi Ono. Many thanks for his voluntary contribution.
References
[1] Akiba, Takuya, et al. “Optuna: A next-generation hyperparameter optimization framework.” Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. ACM, 2019.
[2] Jamil, Momin, and Xin-She Yang. “A literature survey of benchmark functions for global optimization problems.” arXiv preprint arXiv:1308.4008 (2013).
[3] Hansen, Nikolaus, et al. “COCO: A platform for comparing continuous optimizers in a black-box setting.” arXiv preprint arXiv:1603.08785 (2016).
[4] Ying, Chris, et al. “Nas-bench-101: Towards reproducible neural architecture search.” arXiv preprint arXiv:1902.09635 (2019).
[5] Klein, Aaron, and Frank Hutter. “Tabular Benchmarks for Joint Architecture and Hyperparameter Optimization.” arXiv preprint arXiv:1905.04970 (2019).
[6] Dewancker, Ian, et al. “A strategy for ranking optimization methods using multiple criteria.” Workshop on Automatic Machine Learning. 2016.
[7] Bergstra, James S., et al. “Algorithms for hyper-parameter optimization.” Advances in neural information processing systems. 2011.
Tag